2026
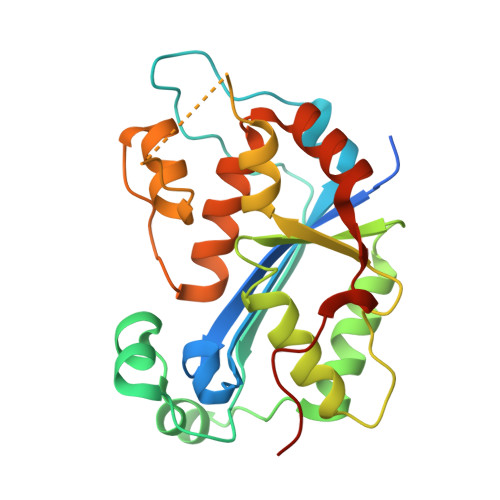
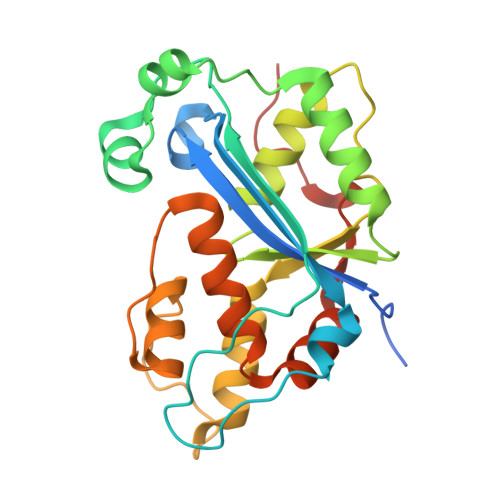
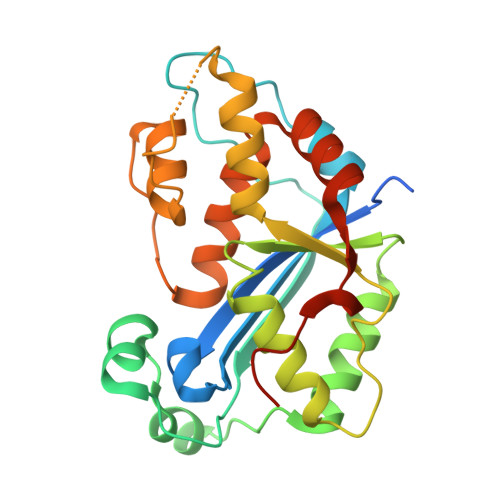
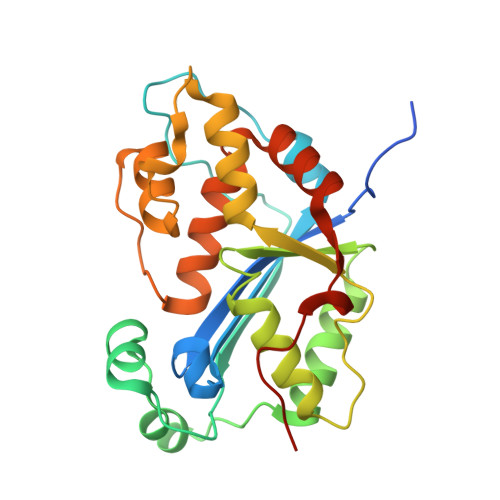
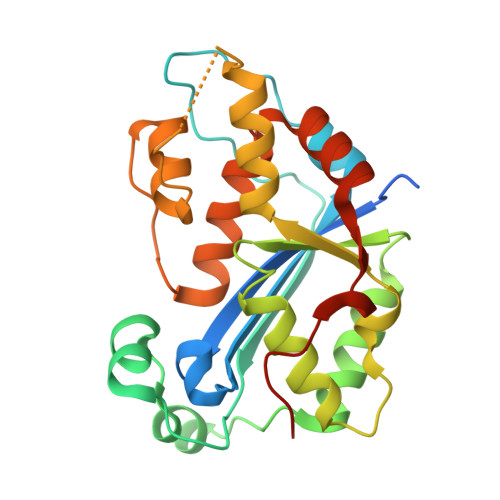
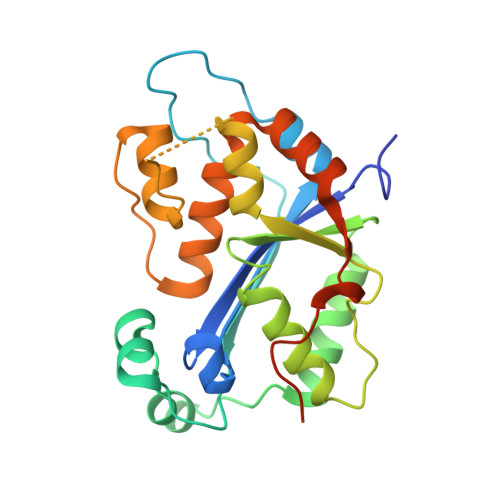
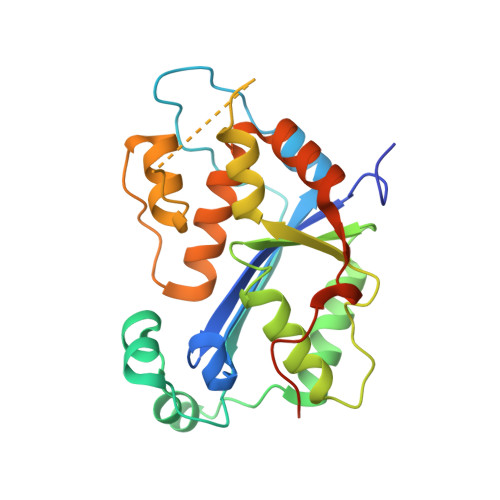
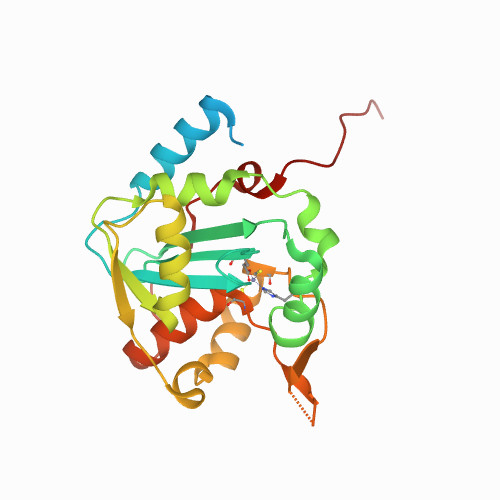
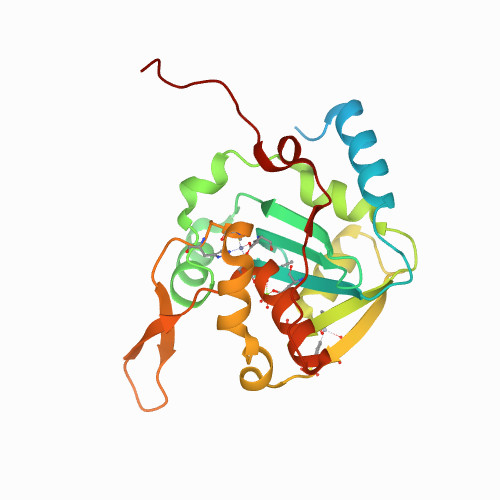
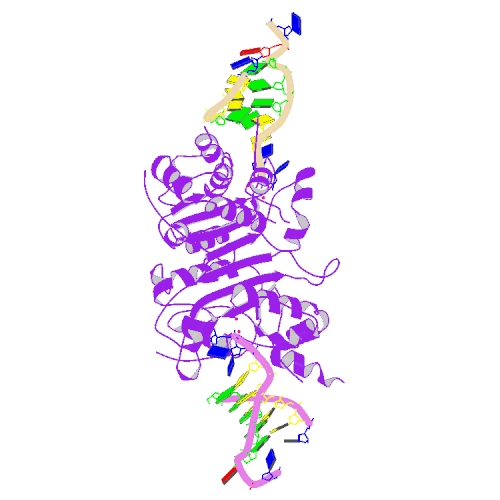
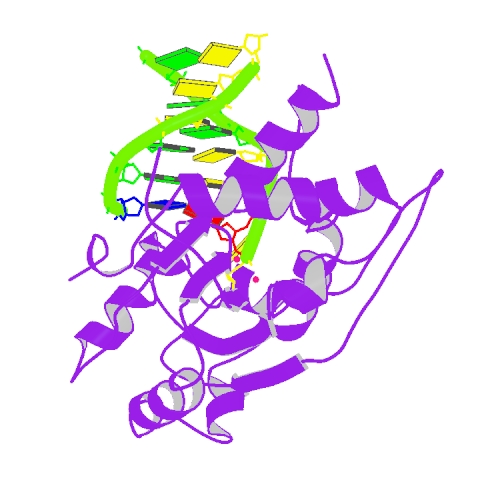
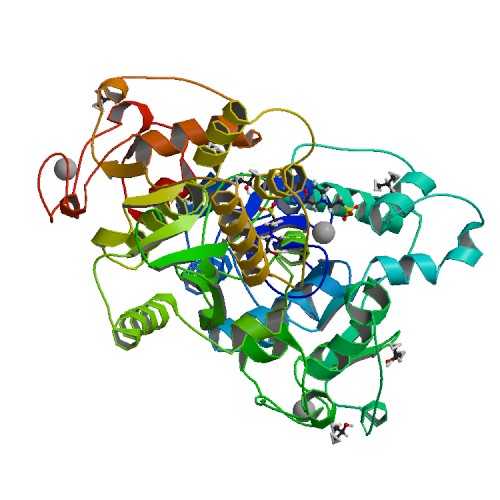
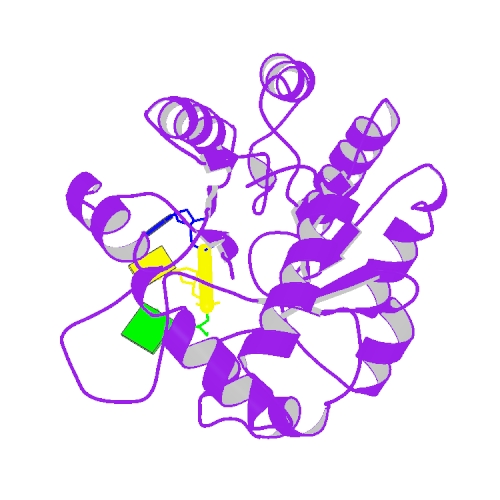
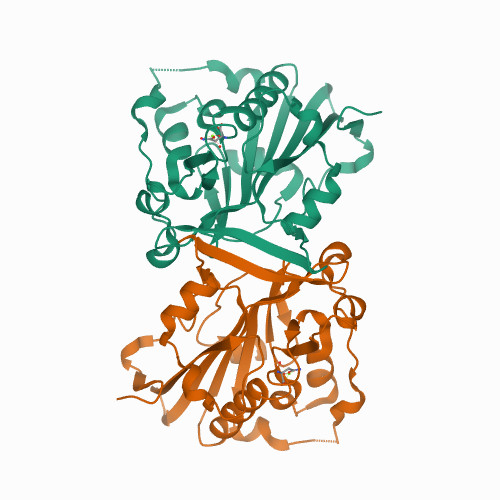
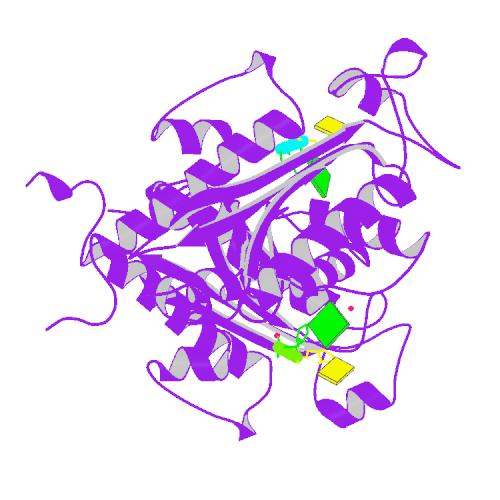
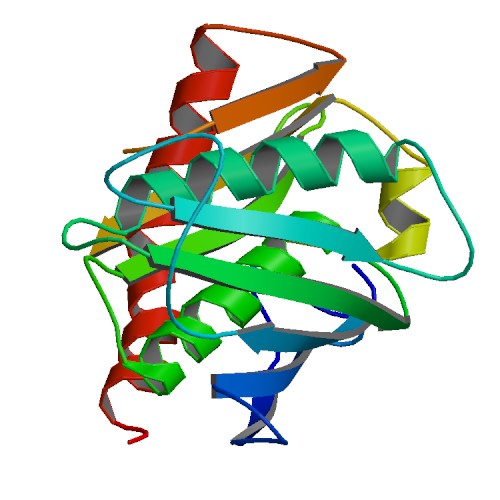
Huang, K.W.; Tsai, C.Y.; Wu, C.Y.; Lin, W.C.; Wu, M.T. ; Hsu, K.C.; Yang, C.Y. ; Chang, I.Y.; Liu, H.M.*; Chu, J.W.*; Hsiao, Y.Y.*, Disordered DNA-binding Motif Forms a Modulation Site for Inhibiting the Cancer Immunotherapy Target TREX1. Nucleic Acids Res., Volume 54, Issue 2, 27 January 2026, https://doi.org/10.1093/nar/gkaf1511 (通訊作者)
Impact factor 2025: 13.1; Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 13/320 (4.06 %)
2023
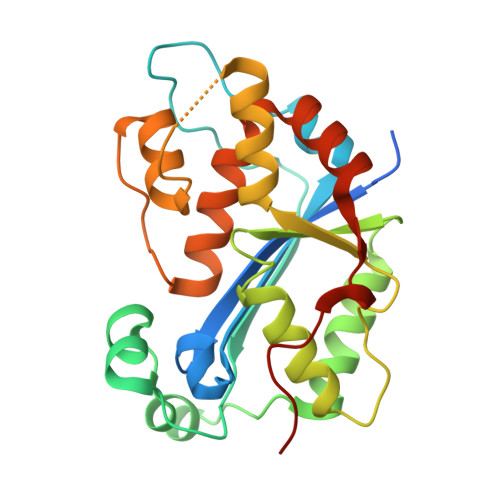
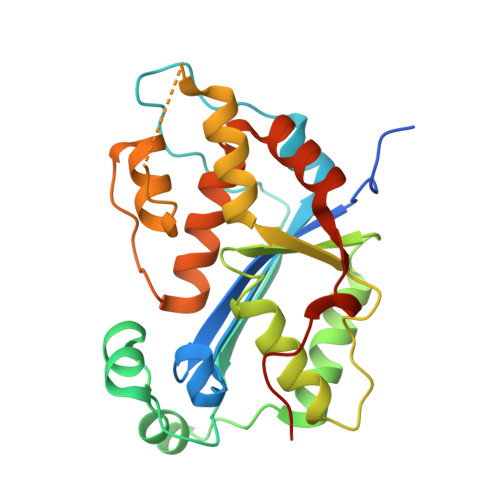
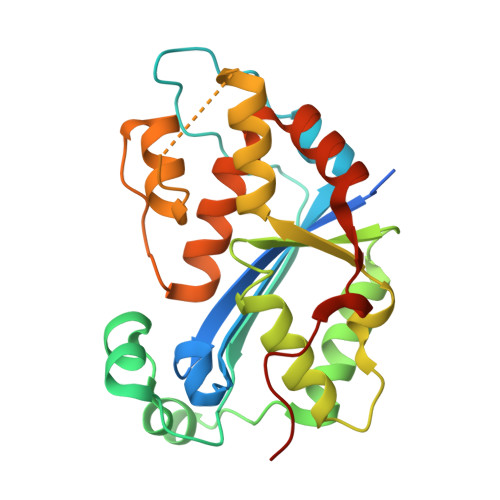
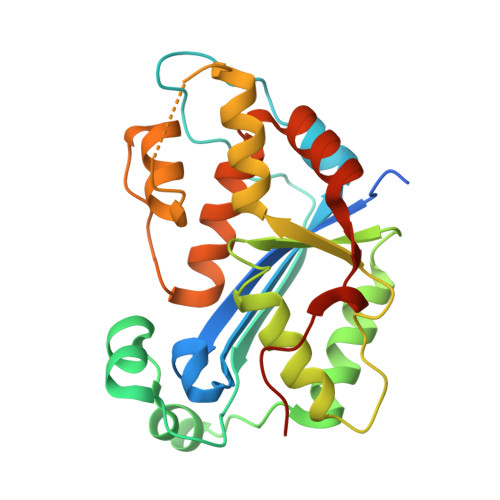
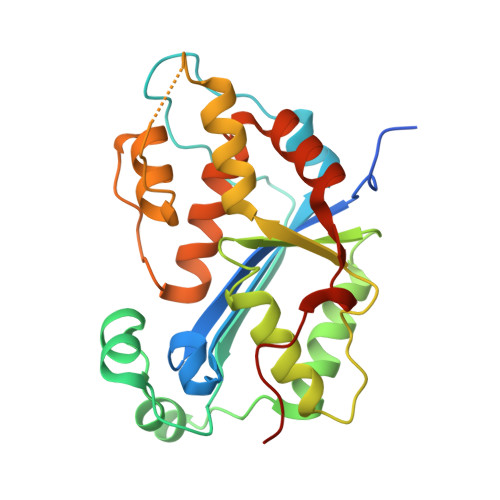
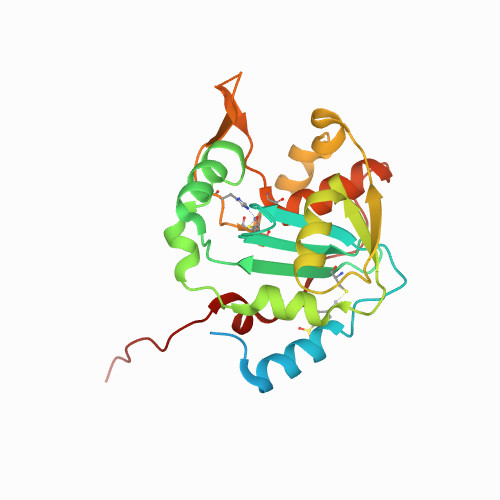
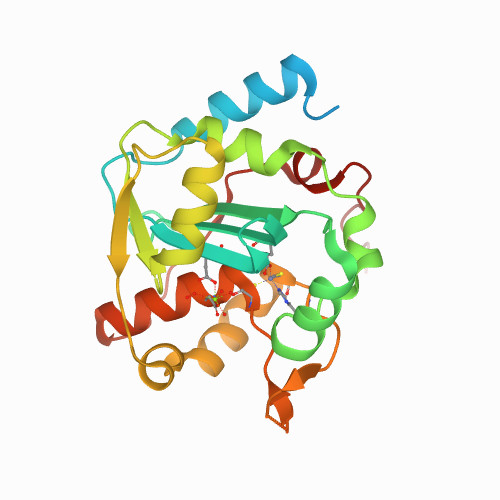
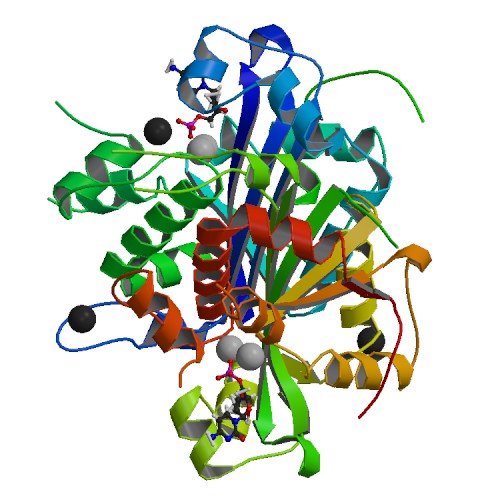
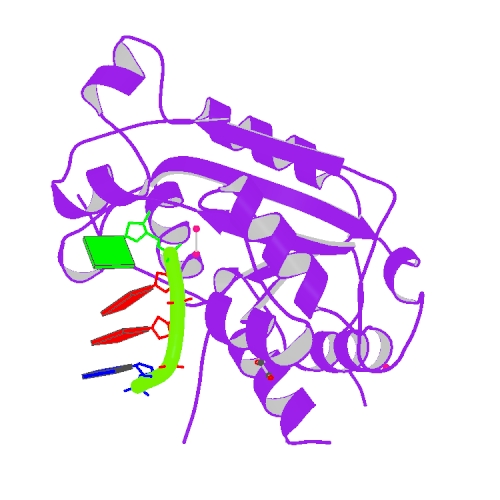
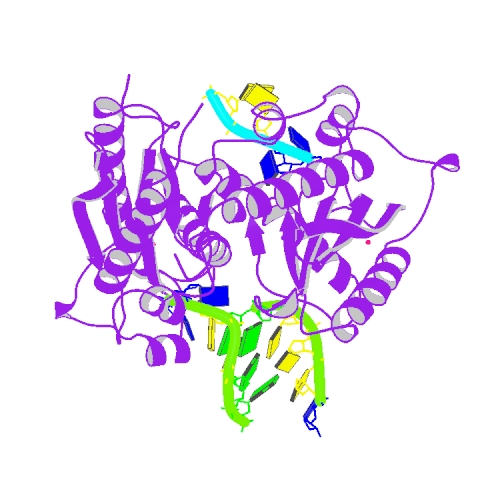
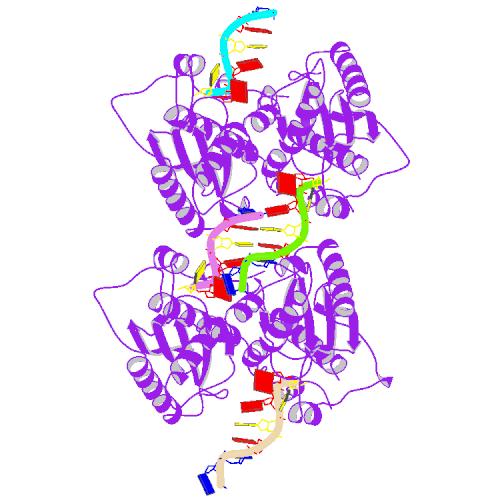
Huang, K.W.; Wu, C.Y.; Toh, S.I.; Liu, T.C.; Tu, C.I; Lin, Y.H.; Cheng, A.J.; Kao, Y.T.; Chu, J.W.*; Hsiao, Y.Y.*, Molecular Insight into the Specific Enzymatic Properties of TREX1 Revealing the Diverse Functions in Processing RNA and DNA/RNA Hybrids. Nucleic Acids Res., Volume 51, Issue 21, 27 November 2023, Pages 11927–11940, https://doi.org/10.1093/nar/gkad910 (通訊作者)
Impact factor 2022: 14.9; Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 10/285 (3.51%)
PDB
2021
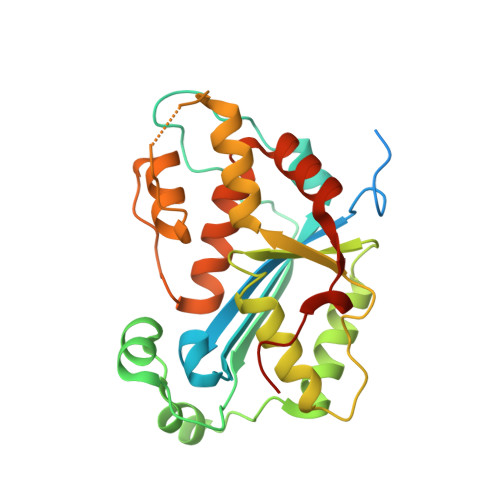
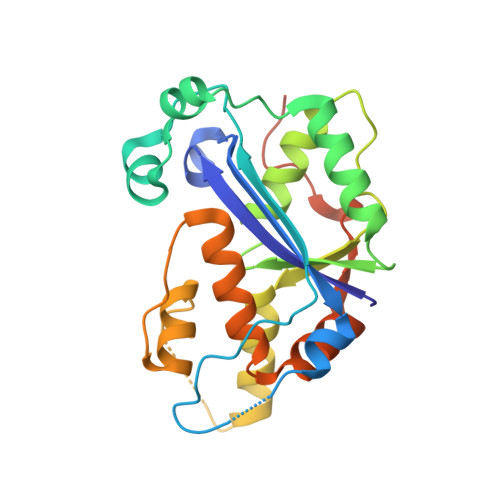
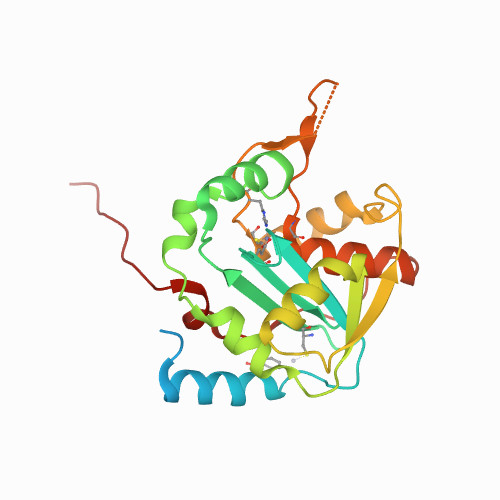
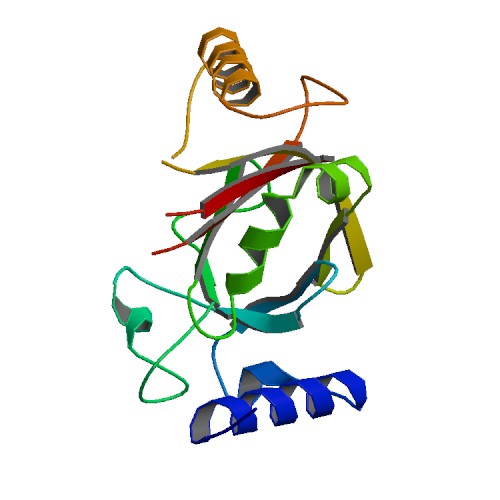
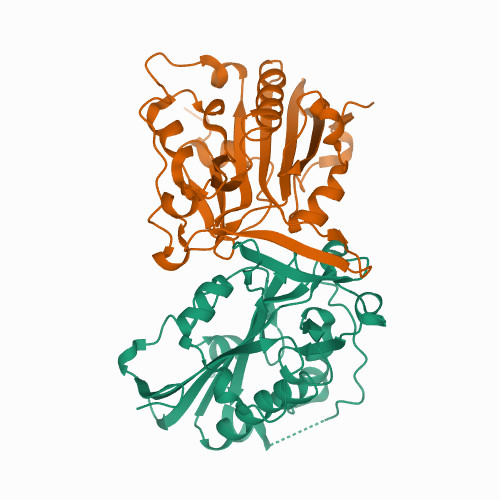
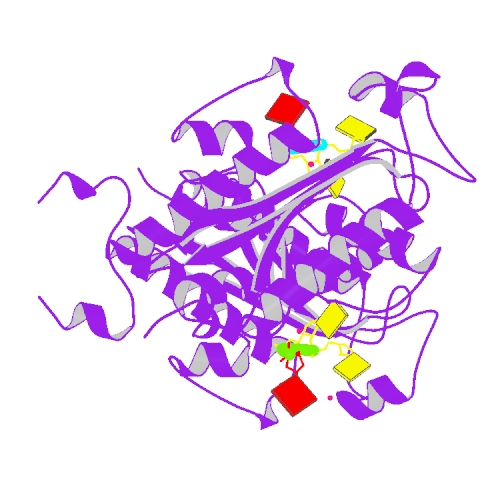
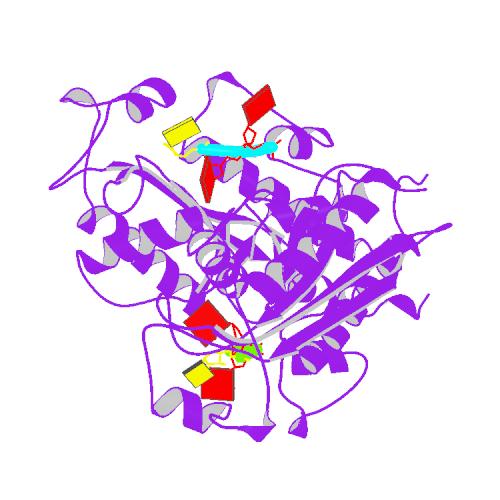
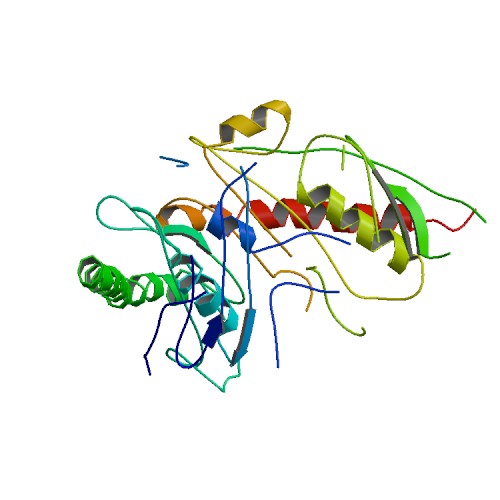
Huang, K.W.; Chen, J.W.; Hua, T.Y.; Chu, Y.Y.; Chiu, T.Y.; Liu, J.Y.; Tu, C.I.; Hsu, K.C.; Kao, Y.T.; Chu, J.W.*; Hsiao, Y.Y.*, Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites. J. Am. Chem. Soc. Au (2021). 1, 12, 2315–2327 ; doi.org/10.1021/jacsau.1c00420 (通訊作者)
Impact factor 2022: 8.0 Ranking: CHEMISTRY, MULTIDISCIPLINARY 27/230 (11.7%)
PDB
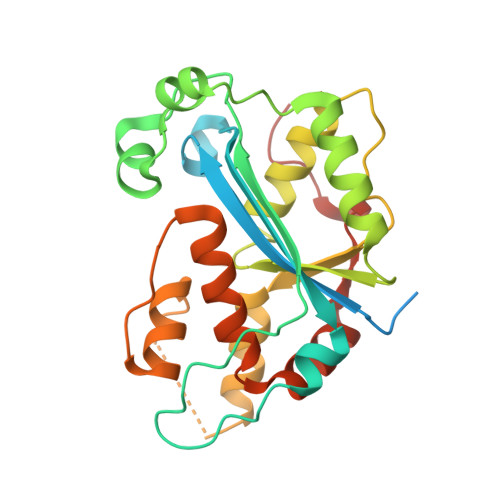
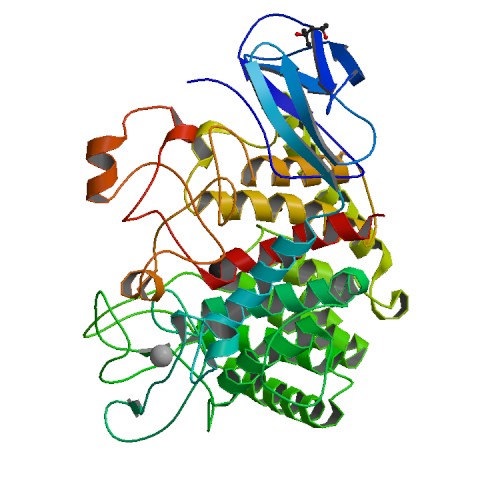
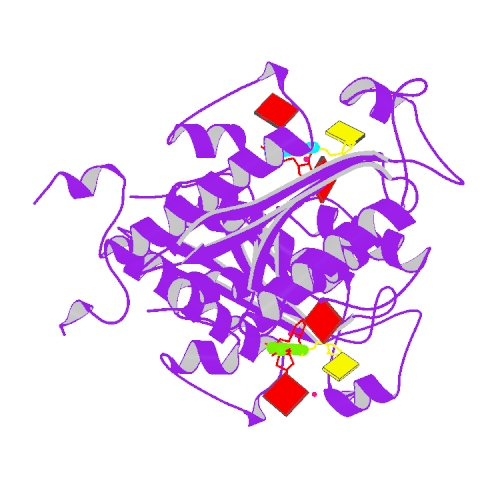
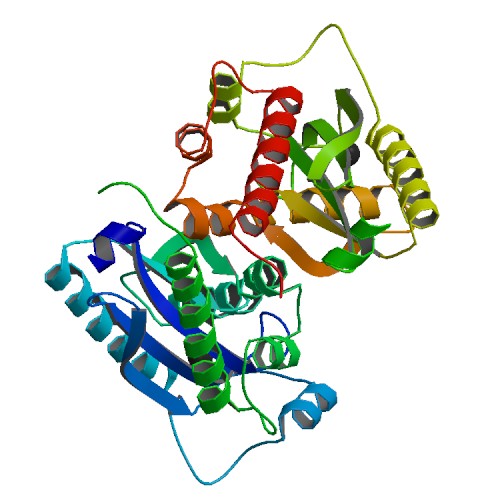
Liu, T.C.; Guo, K.W.; Chu, J.W.*; Hsiao, Y.Y.*, Understanding APE1 Cellular Functions by the Structural Preference of Exonuclease Activities. Comput. Struct. Biotechnol. J. (2021). Jun 24;19:3682-3691. doi: 10.1016/j.csbj.2021.06.036., (通訊作者)
Impact factor 2020: 7.271 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 45/298 (15.0%)
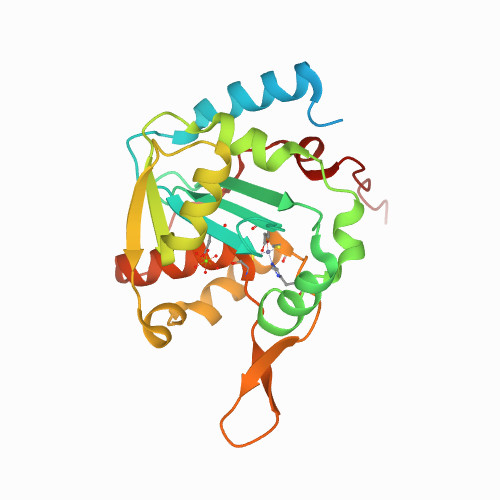
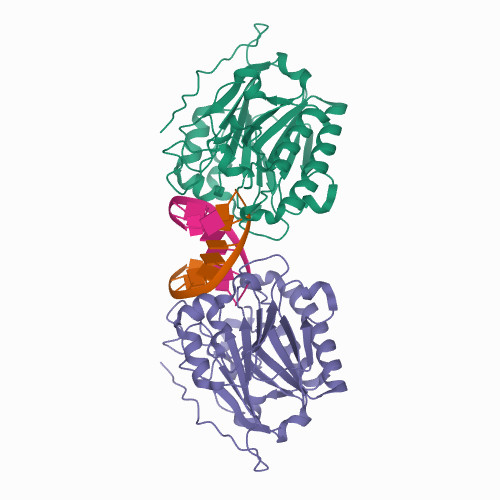
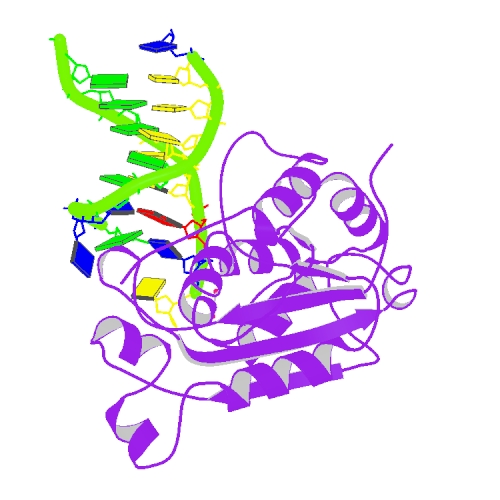
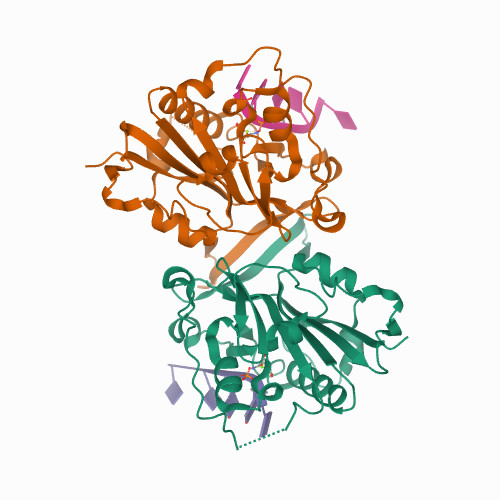
Liu, T.C.; Lin, C.T.; Chang, K.C.; Guo, K.W.; Wang, S.Y.; Chu, J.W.; Hsiao, Y.Y.*, APE1 distinguishes DNA substrates in exonucleolytic cleavage by induced space-filling. Nat Commun. 12, 601 (2021). https://doi.org/10.1038/s41467-020-20853-2. (通訊作者)
Impact factor 2020: 14.919 Ranking: MULTIDISCIPLINARY SCIENCES 4/73 (5.5%)
2020
Kang, C.Y.; Huang, I.H.; Wu, T.Y.; Chang, J.C.; Hsiao, Y.Y.; Cheng, C.H.; Tsai, W.J.; Hsu, K.C.; Wang, S.Y., Functional analysis of Clostridium difficile sortase B reveals key residues for catalytic activity and substrate specificity. J Biol Chem. 2020 Mar 13;295(11):3734-3745. pii: jbc.RA119.011322.
Impact factor 2019: 4.238 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 87/297 (29.1%)
2018
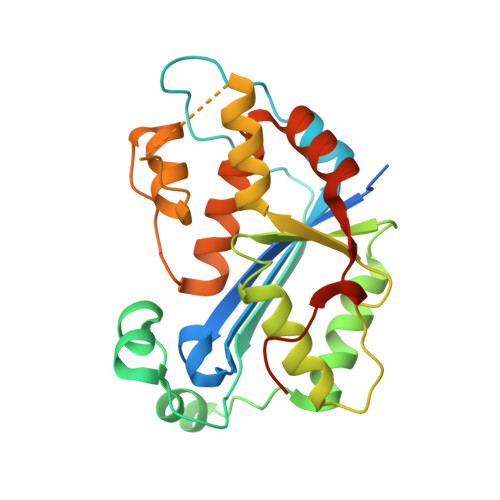
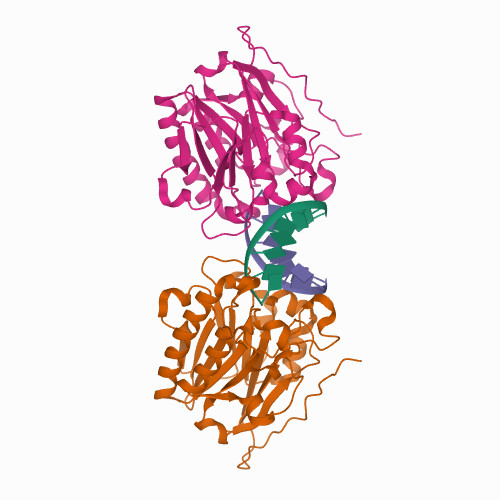
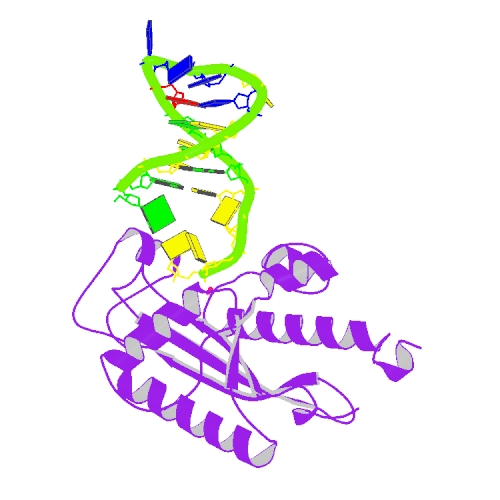
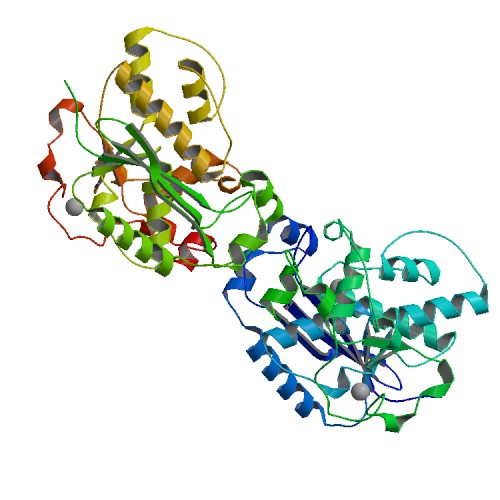
Cheng, H. Lo.; Lin, C.T.; Huang, K.W.; Wang, S.; Lin, Y. T.; Toh, S. I.; Hsiao, Y.Y.*, Structural insights into the duplex DNA processing of TREX2. Nucleic Acids Res.2018 Dec 14;46(22):12166-12176, doi: 10.1093/nar/gky970 (通訊作者)
Impact factor 2017: 11.561 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 10/293 (3.2%)
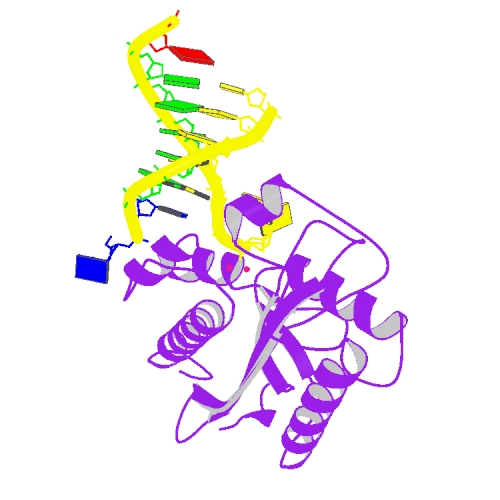
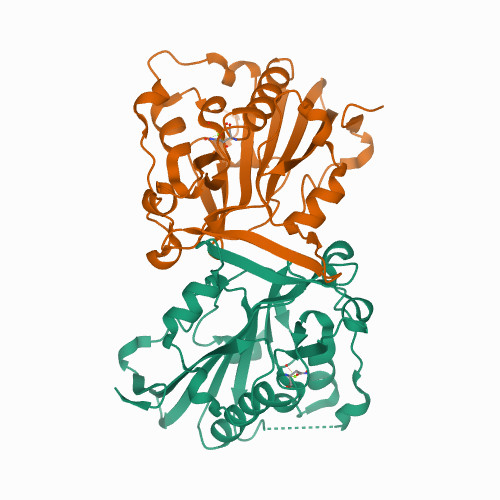
Huang, K. W.; Liu, T. C.; Liang, R. Y.; Chu, L. Y.; Cheng, H. L.; Chu, J. W.; Hsiao, Y.Y.* Structural basis for overhang excision and terminal unwinding of DNA duplexes by TREX1. PLoS Biol. 2018 May 7;16(5):e2005653. doi: 10.1371/journal.pbio.2005653. (通訊作者)
Impact factor 2017: 9.163 Ranking: BIOLOGY 3/85 (3.5%)
2016
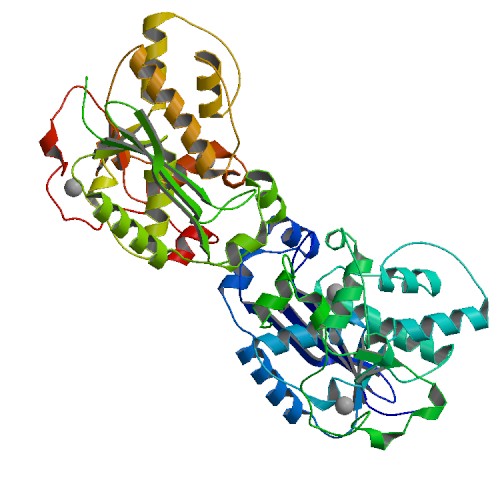
Yin, J. C.; Fei, C. H.; Lo, Y. C.; Hsiao, Y.Y.; Chang, J. C.; Nix, J. C.; Chang, Y. Y.; Yang, L. W.; Huang, I. H.; Wang, S. Structural Insights into Substrate Recognition by Clostridium difficile Sortase. Front Cell Infect Microbiol. 2016, 6, 160.
Impact factor 2015: 5.218 Ranking: MICROBIOLOGY 18/123 (14.2%)
PDB
Wang, H. J.; Hsiao, Y.Y. ; Chen, Y. P.; Ma, T. Y.; Tseng, C. P. Polarity Alteration of a Calcium Site Induces a Hydrophobic Interaction Network and Enhances Cel9A Endoglucanase Thermostability. Appl Environ Microbiol. 2016, 82, 1662-74.
Impact factor 2015: 3.823 Ranking: BIOTECHNOLOGY & APPLIED MICROBIOLOGY 33/161 (20.2%)
PDB
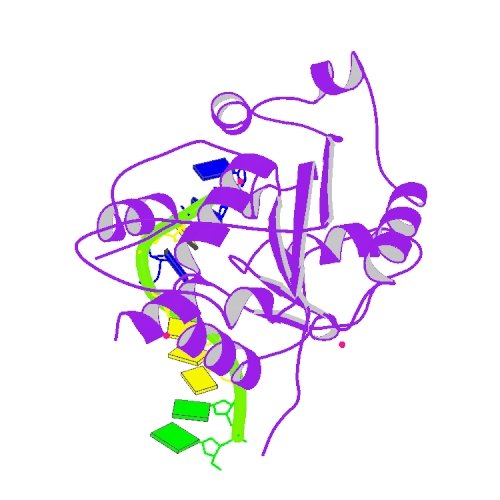
Huang, K. W.; Hsu, K. C.; Chu, L. Y.; Yang, J. M.; Yuan, H. S.*; Hsiao, Y.Y.*, Identification of Inhibitors for the DEDDh Family of Exonucleases and a Unique Inhibition Mechanism by Crystal Structure Analysis of CRN-4 Bound with 2-Morpholin-4-ylethanesulfonate (MES). J Med Chem. 2016, 59, 8019-29. (通訊作者)
Impact factor 2015: 5.589 Ranking: CHEMISTRY, MEDICINAL 3/59 (4.2%)
PDB
2015
Duh, Y.; Hsiao, Y.Y.; Li, C. L.; Huang, J. C.; Yuan, H. S. Aromatic residues in RNase T stack with nucleobases to guide the sequence-specific recognition and cleavage of nucleic acids. Protein Sci. 2015, 24, 1934-41.
Impact factor 2014: 2.854 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 136/290 (46.7%)
2014
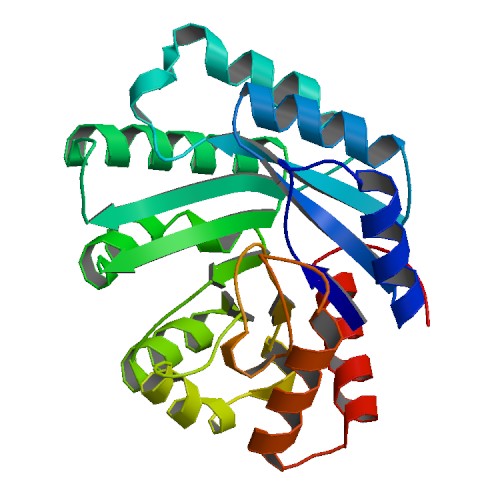
Hsiao, Y.Y.; Fang, W. H.; Lee, C. C.; Chen, Y. P.; Yuan, H. S. Structural insights into DNA repair by RNase T--an exonuclease processing 3' end of structured DNA in repair pathways. PLoS Biol. 2014 Mar 4;12(3):e1001803. doi: 10.1371/journal.pbio.1001803. (第一作者)
Impact factor 2013: 11.771 Ranking: BIOLOGY 1/85 (0.6%)
2012
Lin, J. L., Nakagawa, A., Lin, C. L., Hsiao, Y.Y., Yang, W. Z., Wang, Y. T., Doudeva, L. G., Skeen-Gaar, R. R., Xue, D., & Yuan, H. S. (2012). Structural insights into apoptotic DNA degradation by CED-3 protease suppressor-6 (CPS-6) from Caenorhabditis elegans. J Biol Chem. 287(10), 7110–7120.
Impact factor 2011: 4.773 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 66/290 (22.6%)
Lin, C. L.; Wang, Y. T.; Yang, W. Z.; Hsiao, Y.Y.; Yuan, H. S. Crystal structure of human polynucleotide phosphorylase: insights into its domain function in RNA binding and degradation. Nucleic Acids Res. 2012 May;40(9):4146-57. doi: 10.1093/nar/gkr1281.
Impact factor 2011: 8.026 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 26/290 (8.8%)
Hsiao, Y.Y.; Duh, Y.; Chen, Y. P.; Wang, Y. T.; Yuan, H. S. How an exonuclease decides where to stop in trimming of nucleic acids: crystal structures of RNase T-product complexes. Nucleic Acids Res. 2012, 40, 8144-54. (第一作者)
Impact factor 2011: 8.026 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 26/290 (8.8%)
2011
Hsiao, Y.Y.; Yang, C. C.; Lin, C. L.; Lin, J. L.; Duh, Y.; Yuan, H. S. Structural basis for RNA trimming by RNase T in stable RNA 3'-end maturation. Nat Chem Biol. 2011, 7, 236-43. (第一作者)
Impact factor 2010: 15.808 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 4/286 (1.2%)
2010
Yang, C. C.; Wang, Y. T.; Hsiao, Y.Y.; Doudeva, L. G.; Kuo, P. H.; Chow, S. Y.; Yuan, H. S. Structural and biochemical characterization of CRN-5 and Rrp46: an exosome component participating in apoptotic DNA degradation. RNA. 2010, 16, 1748-59.
Impact factor 2009: 5.198 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 49/283 (17.1%)
2009
Hsiao, Y.Y.; Nakagawa, A.; Shi, Z.; Mitani, S.; Xue, D.; Yuan, H. S. Crystal structure of CRN-4: implications for domain function in apoptotic DNA degradation. Mol Cell Biol. 2009, 29, 448-57. (第一作者)
Impact factor 2008: 5.942 Ranking: BIOCHEMISTRY & MOLECULAR BIOLOGY 38/275 (13.6%)
2008
Hong, T. Y.#; Hsiao, Y.Y.#; Meng, M.; Li, T. T. The 1.5 A structure of endo-1,3-beta-glucanase from Streptomyces sioyaensis: evolution of the active-site structure for 1,3-beta-glucan-binding specificity and hydrolysis. Acta Crystallogr D Biol Crystallogr. 2008, 64, 964-70. (#共同第一作者)
Impact factor 2007: 2.620 Ranking: CRYSTALLOGRAPHY 4/25 (14%)